User:Debra Tabron/sandbox
1,4-Dioxane (14D) does not readily biodegrade in most environments. However, a variety of microorganisms have been identified that can biodegrade 14D through either direct growth-related metabolism or cometabolism. During metabolic biodegradation, microorganisms use 14D as the growth substrate, however growth is slow unless 14D concentrations are very high (>100 mg/L). During cometabolic biodegradation, an additional growth substrate must be supplied to support biomass growth and induce the appropriate 14D-degrading enzymes. Unlike metabolic degradation, cometabolic processes can reduce 14D to very low concentrations.
Related Article(s):
CONTRIBUTOR(S): Dr. Shaily Mahendra and Dr. Michael Hyman
Key Resource(s):
1,4-Dioxane Biodegradation
1,4-Dioxane (14D) does not readily biodegrade in most anaerobic environments[2]. However, multiple studies have demonstrated that 14D can be biodegraded by a variety of microorganisms under aerobic conditions[3][4][5][6]. A summary of microorganisms that are reported to aerobically biodegrade 14D is presented in Table 1.
Under aerobic conditions 14D can be biodegraded through two physiologically distinct processes: (a) metabolism; and (b) cometabolism. Metabolism is a process in which microorganisms use the organic contaminants as a carbon and energy source to support their growth. Cometabolism occurs when microorganisms degrade contaminants using non-specific enzymes but do not gain carbon or energy to support growth from the degradation process.
| Strain | Induced Enzyme | Biodegradation Rate | Reference |
|---|---|---|---|
| Metabolism | |||
| Pseudonocardia dioxanivorans CB1190 | THFMO | 0.19 ± 0.007 mg/hr/mg-protein | Mahendra and Alvarez-Cohen (2005, 2006)[7][3] |
| Actinomycete CB1190* | N/A | 0.33 mg/min/mg-protein | Parales et al. (1994)[8] |
| Amycolata sp. CB1190* | N/A | 0.038 ± 0.012 mg/hr/mg-protein | Kelley et al. (2001) |
| Pseudonocardia benzenivorans B5 | THFMO | 0.01± 0.003 mg/hr/mg-protein | Mahendra and Alvarez-Cohen (2006)[3] |
| Pseudonocardia carboxydivorans RM-31 | ? | 31.6 mg/L/hr | Matsui et al. (2016)[9] |
| Afipia sp. D1 | ? | 0.052 to 0.263 mg/hr/mg-protein | Sei et al. (2013)[10] |
| Mycobacterium sp. PH-06 | PrMO | 2.5 mg/L/hr | Kim et al. (2009)[11] |
| Acinetobacter baumannii DD1 | ? | 2.38 mg/L/hr | Huang et al. (2014)[12] |
| Xanthobacter flavus DT8 | ? | Similar to CB1190 | Chen et al. (2016)[13] |
| Cordyceps sinensis (fungus) | ? | 0.011 mol/day | Nakamiya et al. (2005)[14] |
| Cometabolism | |||
| Mycobacterium austroafricanum JOB5 | ? | 0.40 ± 0.06 mg/hr/mg-protein | House and Hyman (2010)[15] Lan et al. (2013)[16][3] |
| Rhodococcus ruber ENV425 | ? | 10 mg/hr/g TSS | Lippincott et al. (2015)[17]; Vainberg et al. (2006)[18] |
| Pseudonocardia sp. ENV478 | THFMO | 21 mg/hr/g TSS | Masuda et al. (2012)[19] Vainberg et al. (2006)[18] |
| Rhodococcus RR1 | ? | 0.38 ± 0.03 mg/hr/mg-protein | Mahendra and Alvarez-Cohen (2006)[3] |
| Rhodococcus jostii RHA1 | PrMO | N/A | Hand et al. (2015)[20]; Li et al. (2013)[21] |
| Flavobacterium | ? | N/A | Sun et al. (2011)[6] |
| Pseudonocardia K1 | THFMO | 0.26 ± 0.013 mg/hr/mg-protein | Mahendra and Alvarez-Cohen (2006)[3] |
| Burkholderia cepacia G4 | T2MO | 0.1± 0.006 mg/hr/mg-protein | Mahendra and Alvarez-Cohen (2006)[3] |
| Ralstonia pickettii PKO1 | T3MO | 0.31± 0.007 mg/hr/mg-protein | Mahendra and Alvarez-Cohen (2006)[3] |
| Pseudomonas mendocina KR1 | T4MO | 0.37± 0.04 mg/hr/mg-protein | Mahendra and Alvarez-Cohen (2006)[3] |
| Aureobasidium pullmans NRRL 21064 | ? | 6-8 mg/L within a day | Patt and Abebe (1995)[22] |
| Graphium sp. ATCC 58400 (fungus) | CYP | 4 ± 1 nmol/min/mg dry weight (with Propane) 9 ± 5 nmol/min/mg dry weight (with THF) |
Skinner et al. (2009)[23] |
Aerobic Metabolism
While several microorganisms have been isolated that can metabolize 14D[3][11][24][10][12], these organisms are not common. The best characterized 14D-metabolizing strain is Pseudonocardia dioxanivorans CB1190[7]. This bacterium was originally enriched from industrial activated sludge, fed with tetrahydrofuran (THF) and then subsequently fed with 14D[8]. The doubling time of CB1190 was about 30 hours when it was grown in ammonium mineral salts medium at 30 °C amended with 5.5 mM (484 mg/L) 14D[8].
Degradation of 14D by strain CB1190 may be inhibited by elevated concentrations of chlorinated solvents and their degradation products, and by some metals. Inhibition of 14D degradation was strongest for 1,1-dichloroethene (1,1-DCE) followed by cis-1,2-diochloroethene (cDCE) > trichloroethene (TCE) > 1,1,1-trichloroethane (TCA). 14D biodegradation was completely inhibited by 5 mg/L 1,1-DCE[25][20][26]. Cu(II) was the strongest metal inhibitor of 14D degradation by CB1190, causing an increase in the lag period at 1 mg/L and an order of magnitude reduction in 14D degradation rates at 10 and 20 mg/L Cu(II). 14D degradation was less sensitive to Cd(II) and Ni(II), while Zn(II) had no impact on 14D biodegradation at the maximum concentration tested (20 mg/L Zn)[27].
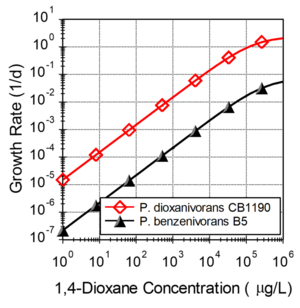
An important challenge for in situ bioremediation of 14D is the slow growth of 14D metabolizers at typical groundwater concentrations. Figure 1 shows estimated growth rates for two 14D metabolizing organisms (P. dioxanivorans CB1190 and P. benzenivorans B5) computed using published kinetic parameters[3]. At a 14D concentrations of 1000 µg/L, CB1190 and B2 have growth rates of 0.015 and 0.002 per day. At typical groundwater concentrations (<1000 µg/L), growth rates are expected to be less than the endogenous decay rate, resulting in a steady decline in the number of 14D metabolizers.
Aerobic Cometabolism
Unlike 14D-metabolizing microorganisms, 14D-cometabolizing organisms do not grow on 14D but can degrade this compound after growth on a primary, growth-supporting substrate. As cometabolically active microorganisms do not use co-substrates as either major carbon or energy sources, they can often degrade co-substrates at concentrations well below those that can be achieved by organisms that metabolize and grow on these co-substrates. A wide variety of bacteria[3] and some fungi[22][23] can cometabolically degrade 14D (see Table 1). These include model organisms that grow on primary substrates such as toluene or methane[3] and involve well-characterized enzymes such as soluble methane monooxygenase (sMMO) or toluene monooxygenases (TxMO), respectively. Other 14D-cometabolizing strains that grow on THF[18], ethane[28], and isobutane[29] have also been described.
Much of the research into 14D cometabolism has focused on high concentrations of 14D (≥100 mg/L), and less is currently known about the activity of specific microorganisms and their monooxygenases at lower, more environmentally relevant 14D concentrations (<100 µg/L). Despite this, many bacterial monooxygenases can concurrently oxidize multiple co-substrates, and recent field studies have demonstrated that cometabolic approaches involving either propane biostimulation and bioaugmentation[17] or propane biostimulation alone[30] can be highly effective treatment strategies for degrading contaminant mixtures that contain parts-per-billion concentrations of both 14D and associated chlorinated co-contaminants.
Anaerobic Biodegradation
To date, there is little evidence of metabolic or cometabolic anaerobic 14D biodegradation. In a microcosm study using samples of aquifer material from several different 14D impacted sites, there was no evidence of 14D biodegradation[31]. However, there is one study that reported anaerobic growth of an iron-reducing bacterium on 14D[2].
An indirect method of anaerobic 14D biodegradation exists via a microbially driven Fenton reaction. Shewanella oneidensis, an Fe(III)-reducing facultative anaerobe, is able to generate hydroxyl radicals that can then break down 14D[32]. Under anaerobic conditions, S. oneidensis produces Fe(II) that can then interact chemically with H2O2 to yield HO• radicals which can oxidatively degrade 1,4-dioxane.
Molecular Biological Tools
Quantitative polymerase chain reaction (qPCR) quantifies the abundance of specific microorganisms and functional genes capable of degrading a particular contaminant. qPCR analyses have been employed to quantify the abundance of microorganisms that can degrade 14D through the activity of tetrahydrofuran monooxygenase (THFMO) in samples from industrial activated sludge and groundwater[33][34][35][21][26]. However, care is warranted in the interpretation of qPCR results in other cases (e.g. propane-stimulated cometabolic 14D degradation) where the role of enzymes such as propane monooxygenase (PrMO) in 14D degradation is less clearly established. Studies also suggest that these biomarkers can be used to detect the abundance of 14D-degraders in situ and how they compete within the larger microbial community[36]. Microbial community analyses can be an asset towards guiding treatment strategies and predicting treatment synergies.
Compound specific isotope analysis (CSIA) refers to measurement of the isotopic signatures of individual chemical compounds and can be used to differentiate contaminant sources, delineate reaction pathways, and provide evidence of in situ contaminant degradation. CSIA methods are now available to measure isotopic fractionation of carbon and hydrogen[29]. For example, the aerobic biodegradation of 14D by a THF-grown 14D-degrading Pseudonocardia strain exhibited an isotopic fractionation factor (ε ) for carbon (εc) of −4.73 ± 0.9‰ and hydrogen (εH) of -147 ± 22‰, respectively. Smaller εcand εH values, (-2.7 ± 0.3‰ and -21 ± 2‰, respectively) were determined for aerobic 14D degradation by a propane-grown Rhodococcus strain. As many Pseudonocardia strains use THFMO to initiate 14D degradation, CSIA, in conjunction with qPCR analyses, may be able to discriminate the roles of metabolism and cometabolism in 14D biodegradation at field sites[37].
Summary
14D is not readily biodegraded under ambient conditions in most environments. However, a variety of microorganisms have been identified that can biodegrade 14D through either growth-related metabolism or fortuitous cometabolism. Metabolic growth on the target pollutant is the more traditional approach for both above ground and in situ bioremediation approaches. However, approaches based on metabolic growth may not be feasible for treatment of 14D at typical concentrations. Cometabolic treatment approaches can reduce 14D to very low levels and have been demonstrated in the field. However, implementation of cometabolic treatment is more complex, and there is currently less engineering experience in the design and operation of these systems.
References
- ^ Zhang, S., Gedalanga, P.B. and Mahendra, S., 2017. Advances in bioremediation of 1, 4-dioxane-contaminated waters. Journal of environmental management, 204, pp.765-774. doi: 10.1016/j.jenvman.2017.05.033
- ^ 2.0 2.1 Shen, W., Chen, H. and Pan, S., 2008. Anaerobic biodegradation of 1, 4-dioxane by sludge enriched with iron-reducing microorganisms. Bioresource technology, 99(7), pp.2483-2487. doi: 10.1016/j.biortech.2007.04.054
- ^ 3.00 3.01 3.02 3.03 3.04 3.05 3.06 3.07 3.08 3.09 3.10 3.11 3.12 Mahendra, S. and Alvarez-Cohen, L., 2006. Kinetics of 1, 4-dioxane biodegradation by monooxygenase-expressing bacteria. Environmental Science & Technology, 40(17), pp.5435-5442. doi:10.1021/es060714v
- ^ Mahendra, S., Petzold, C.J., Baidoo, E.E., Keasling, J.D. and Alvarez-Cohen, L., 2007. Identification of the intermediates of in vivo oxidation of 1, 4-dioxane by monooxygenase-containing bacteria. Environmental Science & Technology, 41(21), pp.7330-7336. doi: 10.1021/es0705745
- ^ Skinner, K., Cuiffetti, L. and Hyman, M., 2009. Metabolism and cometabolism of cyclic ethers by a filamentous fungus, a Graphium sp. Appl. Environ. Microbiol., 75(17), pp.5514-5522. doi:10.1128/AEM.00078-09
- ^ 6.0 6.1 Sun, B., Ko, K. and Ramsay, J.A., 2011. Biodegradation of 1, 4-dioxane by a Flavobacterium. Biodegradation, 22(3), pp.651-659. doi: 10.1007/s10532-010-9438-9
- ^ 7.0 7.1 Mahendra, S. and Alvarez-Cohen, L., 2005. Pseudonocardia dioxanivorans sp. nov., a novel actinomycete that grows on 1, 4-dioxane. International Journal of Systematic and Evolutionary Microbiology, 55(2), pp.593-598. doi: 10.1099/ijs.0.63085-0
- ^ 8.0 8.1 8.2 Parales, R.E., Adamus, J.E., White, N. and May, H.D., 1994. Degradation of 1, 4-dioxane by an actinomycete in pure culture. Applied and Environmental Microbiology, 60(12), pp.4527-4530. Report.pdf
- ^ Matsui, R., Takagi, K., Sakakibara, F., Abe, T. and Shiiba, K., 2016. Identification and characterization of 1, 4-dioxane-degrading microbe separated from surface seawater by the seawater-charcoal perfusion apparatus. Biodegradation, 27(2-3), pp.155-163. [https://doi.org/10.1007/s10532-016-9763-8 doi: 10.1007/s10532-016-9763-8
- ^ 10.0 10.1 Sei, K., Miyagaki, K., Kakinoki, T., Fukugasako, K., Inoue, D. and Ike, M., 2013. Isolation and characterization of bacterial strains that have high ability to degrade 1, 4-dioxane as a sole carbon and energy source. Biodegradation, 24(5), pp.665-674. doi: 10.1007/s10532-012-9614-1
- ^ 11.0 11.1 Kim, Y.M., Jeon, J.R., Murugesan, K., Kim, E.J. and Chang, Y.S., 2009. Biodegradation of 1, 4-dioxane and transformation of related cyclic compounds by a newly isolated Mycobacterium sp. PH-06. Biodegradation, 20(4), p.511. doi: 10.1007/s10532-008-9240-0
- ^ 12.0 12.1 Huang, H., Shen, D., Li, N., Shan, D., Shentu, J. and Zhou, Y., 2014. Biodegradation of 1, 4-dioxane by a novel strain and its biodegradation pathway. Water, Air, & Soil Pollution, 225(9), p.2135. doi: 10.1007/s11270-014-2135-2
- ^ Chen, D.Z., Jin, X.J., Chen, J., Ye, J.X., Jiang, N.X. and Chen, J.M., 2016. Intermediates and substrate interaction of 1, 4-dioxane degradation by the effective metabolizer Xanthobacter flavus DT8. International Biodeterioration & Biodegradation, 106, pp.133-140. doi: 10.1016/j.ibiod.2015.09.018
- ^ Nakamiya, K., Hashimoto, S., Ito, H., Edmonds, J.S. and Morita, M., 2005. Degradation of 1, 4-dioxane and cyclic ethers by an isolated fungus. Appl. Environ. Microbiol., 71(3), pp.1254-1258. doi: 10.1128/AEM.71.3.1254-1258.2005
- ^ House, A.J. and Hyman, M.R., 2010. Effects of gasoline components on MTBE and TBA cometabolism by Mycobacterium austroafricanum JOB5. Biodegradation, 21(4), pp.525-541. doi: 10.1007/s10532-009-9321-8
- ^ Lan, R.S., Smith, C.A. and Hyman, M.R., 2013. Oxidation of cyclic ethers by alkane‐grown Mycobacterium vaccae JOB5. Remediation Journal, 23(4), pp.23-42. [https://doi.org/10.1002/rem.21364 doi: 10.1002/rem.21364}
- ^ 17.0 17.1 Lippincott, D., Streger, S.H., Schaefer, C.E., Hinkle, J., Stormo, J. and Steffan, R.J., 2015. Bioaugmentation and propane biosparging for in situ biodegradation of 1, 4‐dioxane. Groundwater Monitoring & Remediation, 35(2), pp.81-92. doi: 10.1111/gwmr.12093
- ^ 18.0 18.1 18.2 Vainberg, S., McClay, K., Masuda, H., Root, D., Condee, C., Zylstra, G.J. and Steffan, R.J., 2006. Biodegradation of ether pollutants by Pseudonocardia sp. strain ENV478. Appl. Environ. Microbiol., 72(8), pp.5218-5224. doi: 10.1128/AEM.00160-06
- ^ Masuda, H., McClay, K., Steffan, R.J. and Zylstra, G.J., 2012. Biodegradation of tetrahydrofuran and 1, 4-dioxane by soluble diiron monooxygenase in Pseudonocardia sp. strain ENV478. Journal of Molecular Microbiology and Biotechnology, 22(5), pp.312-316. doi: 10.1159/000343817
- ^ 20.0 20.1 Hand, S., Wang, B. and Chu, K.H., 2015. Biodegradation of 1, 4-dioxane: effects of enzyme inducers and trichloroethylene. Science of the Total Environment, 520, pp.154-159. doi: 10.1016/j.scitotenv.2015.03.031
- ^ 21.0 21.1 Li, M., Mathieu, J., Yang, Y., Fiorenza, S., Deng, Y., He, Z., Zhou, J. and Alvarez, P.J., 2013. Widespread distribution of soluble di-iron monooxygenase (SDIMO) genes in arctic groundwater impacted by 1, 4-dioxane. Environmental science & technology, 47(17), pp.9950-9958. doi: 10.1021/es402228x
- ^ 22.0 22.1 Patt, T.E. and Abebe, H.M., Upjohn Co, 1995. Microbial degradation of chemical pollutants. U.S. Patent 5,399,495. Report.pdf
- ^ 23.0 23.1 Skinner, K., Cuiffetti, L. and Hyman, M., 2009. Metabolism and cometabolism of cyclic ethers by a filamentous fungus, a Graphium sp. Appl. Environ. Microbiol., 75(17), pp.5514-5522. doi:10.1128/AEM.00078-09
- ^ Sei, K., Kakinoki, T., Inoue, D., Soda, S., Fujita, M. and Ike, M., 2010. Evaluation of the biodegradation potential of 1, 4-dioxane in river, soil and activated sludge samples. Biodegradation, 21(4), pp.585-591. doi: 10.1007/s10532-010-9326-3
- ^ Mahendra, S., Grostern, A. and Alvarez-Cohen, L., 2013. The impact of chlorinated solvent co-contaminants on the biodegradation kinetics of 1, 4-dioxane. Chemosphere, 91(1), pp.88-92. doi: 10.1016/j.chemosphere.2012.10.104
- ^ 26.0 26.1 Zhang, S., Gedalanga, P.B. and Mahendra, S., 2016. Biodegradation kinetics of 1, 4-dioxane in chlorinated solvent mixtures. Environmental Science & Technology, 50(17), pp.9599-9607. doi: 10.1021/acs.est.6b02797
- ^ Pornwongthong, P., Mulchandani, A., Gedalanga, P.B. and Mahendra, S., 2014. Transition metals and organic ligands influence biodegradation of 1, 4-dioxane. Applied biochemistry and biotechnology, 173(1), pp.291-306. doi: 10.1007/s12010-014-0841-2
- ^ Hatzinger, P.B., Banerjee, R., Rezes, R., Streger, S.H., McClay, K. and Schaefer, C.E., 2017. Potential for cometabolic biodegradation of 1, 4-dioxane in aquifers with methane or ethane as primary substrates. Biodegradation, 28(5-6), pp.453-468. doi: 0.1007/s10532-017-9808-7
- ^ 29.0 29.1 Bennett, P., Hyman, M., Smith, C., El Mugammar, H., Chu, M.Y., Nickelsen, M. and Aravena, R., 2018. Enrichment with carbon-13 and deuterium during monooxygenase-mediated biodegradation of 1, 4-dioxane. Environmental Science & Technology Letters, 5(3), pp.148-153. doi: 10.1021/acs.estlett.7b00565
- ^ Chu, M.Y.J., Bennett, P.J., Dolan, M.E., Hyman, M.R., Peacock, A.D., Bodour, A., Anderson, R.H., Mackay, D.M. and Goltz, M.N., 2018. Concurrent Treatment of 1, 4‐Dioxane and Chlorinated Aliphatics in a Groundwater Recirculation System Via Aerobic Cometabolism. Groundwater Monitoring & Remediation, 38(3), pp.53-64. doi: 10.1111/gwmr.12293
- ^ Zenker, M.J., Borden, R.C. and Barlaz, M.A., 1999. Investigation of the intrinsic biodegradation of alkyl and cyclic ethers. The Fifth International In Situ and On-Site Bioremediation Symposium
- ^ Sekar, R. and DiChristina, T.J., 2014. Microbially driven Fenton reaction for degradation of the widespread environmental contaminant 1, 4-dioxane. Environmental science & technology, 48(21), pp.12858-12867. doi: 10.1021/es503454a
- ^ Chiang, S.Y.D., Mora, R., Diguiseppi, W.H., Davis, G., Sublette, K., Gedalanga, P. and Mahendra, S., 2012. Characterizing the intrinsic bioremediation potential of 1, 4-dioxane and trichloroethene using innovative environmental diagnostic tools. Journal of Environmental Monitoring, 14(9), pp.2317-2326. doi: 10.1039/C2EM30358B
- ^ Gedalanga, P.B., Pornwongthong, P., Mora, R., Chiang, S.Y.D., Baldwin, B., Ogles, D. and Mahendra, S., 2014. Identification of biomarker genes to predict biodegradation of 1, 4-dioxane. Appl. Environ. Microbiol., 80(10), pp.3209-3218. doi: 10.1128/AEM.04162-13
- ^ Li, M., Mathieu, J., Liu, Y., Van Orden, E.T., Yang, Y., Fiorenza, S. and Alvarez, P.J., 2013. The abundance of tetrahydrofuran/dioxane monooxygenase genes (thmA/dxmA) and 1, 4-dioxane degradation activity are significantly correlated at various impacted aquifers. Environmental Science & Technology Letters, 1(1), pp.122-127. doi: 10.1021/ez400176h
- ^ Miao, Y., Johnson, N.W., Gedalanga, P.B., Adamson, D., Newell, C. and Mahendra, S., 2019. Response and recovery of microbial communities subjected to oxidative and biological treatments of 1, 4-dioxane and co-contaminants. Water research, 149, pp.74-85. doi:10.1016/j.watres.2018.10.070
- ^ Gedalanga, P., Madison, A., Miao, Y., Richards, T., Hatton, J., DiGuiseppi, W.H., Wilson, J. and Mahendra, S., 2016. A Multiple Lines of Evidence Framework to Evaluate Intrinsic Biodegradation of 1, 4‐Dioxane. Remediation Journal, 27(1), pp.93-114. doi: 10.1002/rem.21499