Difference between revisions of "Stable Isotope Probing (SIP)"
| (43 intermediate revisions by 2 users not shown) | |||
| Line 3: | Line 3: | ||
'''Related Article(s):''' | '''Related Article(s):''' | ||
| + | |||
| + | *[[Compound Specific Isotope Analysis (CSIA)]] | ||
| + | *[[Metagenomics]] | ||
*[[Molecular Biological Tools - MBTs]] | *[[Molecular Biological Tools - MBTs]] | ||
*[[Quantitative Polymerase Chain Reaction (qPCR)]] | *[[Quantitative Polymerase Chain Reaction (qPCR)]] | ||
| − | |||
| − | |||
| − | ''' | + | '''Contributor(s):''' [[Dora Ogles-Taggart]] and [[Dr. Brett Baldwin]] |
'''Key Resource(s):''' | '''Key Resource(s):''' | ||
| − | *[http://dx.doi.org/10.1007/s10532-011-9455-3 Identification of TBA-utilizing organisms in BioGAC reactors using <sup>13</sup>C-DNA SIP]<ref name ="Aslett2011"> Aslett, D., Haas, J., Hyman, M., 2011. Identification of tertiary butyl alcohol(TBA)-utilizing organisms in BioGAC reactors using <sup>13</sup>C-DNA stable isotope probing. Biodegradation, 22(5), 961-972. [http://dx.doi.org/10.1007/s10532-011-9455-3 doi:10.1007/s10532-011-9455-3]</ref> | + | |
| − | *[http://onlinelibrary.wiley.com/doi/10.1111/gwmr.12031/abstract Using DNA‐Stable Isotope Probing to Identify MTBE‐and TBA‐Degrading Microorganisms in Contaminated Groundwater]<ref name = "Key2013">Key, K.C., Sublette, K.L., Duncan, K., Mackay, D.M., Scow, K.M., Ogles, D., 2013. Using DNA‐Stable Isotope Probing to Identify MTBE‐and TBA‐Degrading Microorganisms in Contaminated Groundwater. Groundwater Monitoring & Remediation, 33(4), 57-68. [http://onlinelibrary.wiley.com/doi/10.1111/gwmr.12031/abstract doi:10.1111/gwmr.12031]</ref> | + | *[http://dx.doi.org/10.1007/s10532-011-9455-3 Identification of TBA-utilizing organisms in BioGAC reactors using <sup>13</sup>C-DNA SIP]<ref name="Aslett2011"> Aslett, D., Haas, J., Hyman, M., 2011. Identification of tertiary butyl alcohol(TBA)-utilizing organisms in BioGAC reactors using <sup>13</sup>C-DNA stable isotope probing. Biodegradation, 22(5), 961-972. [http://dx.doi.org/10.1007/s10532-011-9455-3 doi:10.1007/s10532-011-9455-3]</ref> |
| + | *[http://onlinelibrary.wiley.com/doi/10.1111/gwmr.12031/abstract Using DNA‐Stable Isotope Probing to Identify MTBE‐and TBA‐Degrading Microorganisms in Contaminated Groundwater]<ref name="Key2013">Key, K.C., Sublette, K.L., Duncan, K., Mackay, D.M., Scow, K.M., Ogles, D., 2013. Using DNA‐Stable Isotope Probing to Identify MTBE‐and TBA‐Degrading Microorganisms in Contaminated Groundwater. Groundwater Monitoring & Remediation, 33(4), 57-68. [http://onlinelibrary.wiley.com/doi/10.1111/gwmr.12031/abstract doi:10.1111/gwmr.12031]</ref> | ||
==Introduction== | ==Introduction== | ||
Stable isotope probing (SIP) is used to conclusively determine whether in situ biodegradation of a contaminant is occurring. The “probe” is a synthesized version of the contaminant compound composed of the heavier stable isotope (e.g., <sup>13</sup>C, <sup>15</sup>N) rather than the more common light isotope of that element (e.g., <sup>12</sup>C, <sup>14</sup>N). The heavy isotope serves as the “label” to track the environmental fate of the contaminant and determine if biodegradation is occurring. | Stable isotope probing (SIP) is used to conclusively determine whether in situ biodegradation of a contaminant is occurring. The “probe” is a synthesized version of the contaminant compound composed of the heavier stable isotope (e.g., <sup>13</sup>C, <sup>15</sup>N) rather than the more common light isotope of that element (e.g., <sup>12</sup>C, <sup>14</sup>N). The heavy isotope serves as the “label” to track the environmental fate of the contaminant and determine if biodegradation is occurring. | ||
| − | SIP is used to conclusively determine if biodegradation of some common contaminants like petroleum hydrocarbons (e.g., [[wikipedia: BTX (chemistry) | BTEX]], [[Polycyclic Aromatic Hydrocarbons (PAHs) | polycyclic aromatic hydrocarbons (PAHs)]]) and oxygenates (e.g., [[wikipedia: Methyl tert-butyl ether | MTBE]], [[wikipedia: tert-Butyl alcohol | TBA]]) is occurring. Results are often used to evaluate the feasibility of [[Monitored Natural Attenuation (MNA)| monitored natural attenuation (MNA)]] as a site management strategy<ref>Busch‐Harris, J., Sublette, K., Roberts, K.P., Landrum, C., Peacock, A.D., Davis, G., Ogles, D., Holmes, W.E., Harris, D., Ota, C., Yang, X., 2008. Bio‐Traps Coupled with Molecular Biological Methods and Stable Isotope Probing Demonstrate the In | + | SIP is used to conclusively determine if biodegradation of some common contaminants like petroleum hydrocarbons (e.g., [[wikipedia: BTX (chemistry) | BTEX]], [[Polycyclic Aromatic Hydrocarbons (PAHs) | polycyclic aromatic hydrocarbons (PAHs)]]) and oxygenates (e.g., [[wikipedia: Methyl tert-butyl ether | MTBE]], [[wikipedia: tert-Butyl alcohol | TBA]]) is occurring. Results are often used to evaluate the feasibility of [[Monitored Natural Attenuation (MNA)| monitored natural attenuation (MNA)]] as a site management strategy<ref>Busch‐Harris, J., Sublette, K., Roberts, K.P., Landrum, C., Peacock, A.D., Davis, G., Ogles, D., Holmes, W.E., Harris, D., Ota, C., Yang, X., 2008. Bio‐Traps Coupled with Molecular Biological Methods and Stable Isotope Probing Demonstrate the In Situ Biodegradation Potential of MTBE and TBA in Gasoline‐Contaminated Aquifers. Groundwater Monitoring & Remediation, 28(4), 47-62. [http://dx.doi.org/10.1111/j.1745-6592.2008.00216.x doi:10.1111/j.1745-6592.2008.00216.x]</ref><ref>Geyer, R., Peacock, A.D., Miltner, A., Richnow, H.H., White, D.C., Sublette, K.L., Kästner, M., 2005. In situ assessment of biodegradation potential using biotraps amended with <sup>13</sup>C-labeled benzene or toluene. Environmental Science & Technology, 39(13), 4983-4989. [http://dx.doi.org/10.1021/es048037x doi:10.1021/es048037x]</ref><ref>Key, K.C., Sublette, K.L., Johannes, T.W., Ogles, D., Baldwin, B., Biernacki, A., 2014. Assessing BTEX Biodegradation Potential at a Refinery Using Molecular Biological Tools. Groundwater Monitoring & Remediation, 34(1), 35-48. [http://onlinelibrary.wiley.com/doi/10.1111/gwmr.12037/abstract doi:10.1111/gwmr.12037]</ref><ref>Williams, N., Hyland, A., Mitchener, R., Sublette, K., Key, K.C., Davis, G., Ogles, D., Baldwin, B., Biernacki, A., 2013. Demonstrating the In Situ Biodegradation Potential of Phenol Using Bio‐Sep<sup>®</sup> Bio‐Traps<sup>®</sup> and Stable Isotope Probing. Remediation Journal, 23(1), 7-22. [http://onlinelibrary.wiley.com/doi/10.1002/rem.21335/abstract doi:10.1002/rem.21335]</ref>. Conclusive evidence of contaminant biodegradation increases stakeholder confidence that MNA is more than a “do nothing” alternative. While often performed to assess MNA, SIP can also be used to evaluate the feasibility and performance of [[Remediation Technologies | engineered bioremediation]] approaches. In research settings, SIP can also be used with DNA-based analyses to help identify the organisms involved in specific biodegradation processes<ref name="Aslett2011" /><ref name="Hatzinger2014">Hatzinger, P., Fuller, M., 2014. New approaches to evaluate the biological degradation of RDX in groundwater. Project ER-1607. Strategic Environmental Research Development Program, Arlington, VA. [https://www.serdp-estcp.org/Program-Areas/Environmental-Restoration/Contaminants-on-Ranges/Characterizing-Fate-and-Transport/ER-1607 ER-1607]</ref><ref name="Key2013" />. |
==How Does SIP Work== | ==How Does SIP Work== | ||
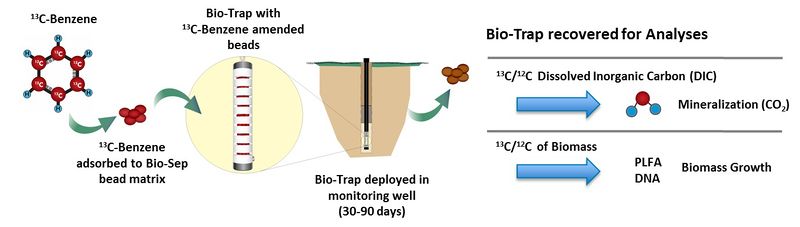
| − | In a typical SIP study, a <sup>13</sup>C form of the contaminant (e.g., <sup>13</sup>C-benzene) is adsorbed to a [[No-Purge/Passive | + | In a typical SIP study, a <sup>13</sup>C form of the contaminant (e.g., <sup>13</sup>C-benzene) is adsorbed to a [[Groundwater Sampling - No-Purge/Passive | passive microbial sampling device]] such as Bio-Sep<sup>®</sup> beads inside a Bio-Trap<sup>®</sup> sampler. Bio-Sep<sup>®</sup> beads are an engineered composite of Nomex<sup>®</sup> and powdered activated carbon (PAC). PAC adsorbs the <sup>13</sup>C-labeled compound and also provides a large surface area for microbial colonization and growth. Nomex<sup>®</sup> allows the beads to be heat sterilized prior to the study. Such a passive sampler is deployed in a monitoring well (Fig. 1). |
[[File:Fig1 SIP Olges.jpg|thumbnail|left|800px|Figure 1. SIP in use: Loading, deployment and recovery of Bio-Trap<sup>®</sup> passive sampler with <sup>13</sup>C-labeled benzene.]] | [[File:Fig1 SIP Olges.jpg|thumbnail|left|800px|Figure 1. SIP in use: Loading, deployment and recovery of Bio-Trap<sup>®</sup> passive sampler with <sup>13</sup>C-labeled benzene.]] | ||
| Line 31: | Line 33: | ||
==Advantages== | ==Advantages== | ||
| + | |||
*'''Conclusive''': SIP can provide conclusive evidence that in situ biodegradation of the contaminant is occurring. | *'''Conclusive''': SIP can provide conclusive evidence that in situ biodegradation of the contaminant is occurring. | ||
| Line 40: | Line 43: | ||
==Limitations== | ==Limitations== | ||
| − | *'''Not applicable to all contaminants''': SIP is generally not appropriate for compounds that are used as terminal electron acceptors, such as [[wikipedia: Trichloroethylene | trichloroethylene (TCE)]] and other chlorinated ethenes, because the <sup>13</sup>C label is not incorporated into biomass or CO<sub>2 </sub> during this microbial process. Tools such as [[Compound Specific Isotope Analysis (CSIA) | + | |
| + | *'''Not applicable to all contaminants''': SIP is generally not appropriate for compounds that are used as terminal electron acceptors, such as [[wikipedia: Trichloroethylene | trichloroethylene (TCE)]] and other chlorinated ethenes, because the <sup>13</sup>C label is not incorporated into biomass or CO<sub>2 </sub> during this microbial process. Tools such as [[Compound Specific Isotope Analysis (CSIA) | compound-specific isotope analysis (CSIA)]] performed on the contaminant itself is more applicable for these compounds. For large or more complex compounds, synthesis of the <sup>13</sup>C-labeled compound can be expensive or simply not available. | ||
*'''Dilute plumes''': Data obtained from typical SIP studies where concentrations of <sup>13</sup>C-labeled compounds are relatively high may not necessarily extrapolate to biodegradation of the contaminant present in a dilute plume where contaminant concentrations are approaching closure levels. | *'''Dilute plumes''': Data obtained from typical SIP studies where concentrations of <sup>13</sup>C-labeled compounds are relatively high may not necessarily extrapolate to biodegradation of the contaminant present in a dilute plume where contaminant concentrations are approaching closure levels. | ||
==Reporting SIP Results== | ==Reporting SIP Results== | ||
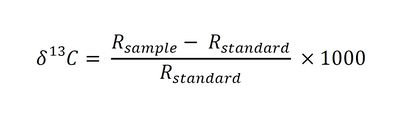
| − | SIP results are reported as <sup>13</sup>C/<sup>12</sup>C ratios in delta notation (δ<sup>13</sup>C) with units of parts per thousand (“per mil” ‰), where δ<sup>13</sup>C is defined as follows<ref name="LABSIE">Laboratory S.I.E. Overview of Stable Isotope Research. | + | SIP results are reported as <sup>13</sup>C/<sup>12</sup>C ratios in delta notation (δ<sup>13</sup>C) with units of parts per thousand (“per mil” ‰), where δ<sup>13</sup>C is defined as follows<ref name="LABSIE">Laboratory S.I.E. Overview of Stable Isotope Research. http://sisbl.uga.edu/stable.html</ref><ref>USGS Fundamentals of Stable Isotope Geochemistry. [http://wwwrcamnl.wr.usgs.gov/isoig/res/funda.html USGS Resources on Isotopes]</ref>: |
[[File:Ogles equ.JPG|center|400px]] | [[File:Ogles equ.JPG|center|400px]] | ||
| − | ::Where: | + | |
| − | ::R<sub>sample</sub> is the <sup>13</sup>C/<sup>12</sup>C ratio of the sample and | + | ::Where: |
| + | ::R<sub>sample</sub> is the <sup>13</sup>C/<sup>12</sup>C ratio of the sample and | ||
::R<sub>standard</sub> is the <sup>13</sup>C/<sup>12</sup>C ratio of the Vienna Pee Dee Belemnite (VPDB) standard. | ::R<sub>standard</sub> is the <sup>13</sup>C/<sup>12</sup>C ratio of the Vienna Pee Dee Belemnite (VPDB) standard. | ||
==Background δ<sup>13</sup>C Values== | ==Background δ<sup>13</sup>C Values== | ||
| − | The <sup>13</sup>C/<sup>12</sup>C ratio of the VPDB standard (1.1112328% <sup>13</sup>C) is greater than most other natural carbon-based substances<ref name="LABSIE"/>. Because R<sub>standard</sub> is typically larger than R<sub>sample </sub>, calculated δ<sup>13</sup>C values of most naturally-occurring materials will be negative values (0‰ to -110‰). | + | The <sup>13</sup>C/<sup>12</sup>C ratio of the VPDB standard (1.1112328% <sup>13</sup>C) is greater than most other natural carbon-based substances<ref name="LABSIE" />. Because R<sub>standard</sub> is typically larger than R<sub>sample </sub>, calculated δ<sup>13</sup>C values of most naturally-occurring materials will be negative values (0‰ to -110‰). |
The isotopic composition of subsurface microorganisms and their metabolic products (e.g., CO<sub>2 </sub>) will directly reflect the isotopic composition of the compounds used as carbon and energy sources. Thus under natural conditions, background δ<sup>13</sup>C values for microbial biomass and DIC would be expected to be negative and are often between -20‰ and -30‰ <ref>Pelz, O., Chatzinotas, A., Zarda-Hess, A., Abraham, W.R., Zeyer, J., 2001. Tracing toluene-assimilating sulfate-reducing bacteria using <sup>13</sup>C-incorporation in fatty acids and whole-cell hybridization. FEMS Microbiology Ecology, 38(2-3), 123-131. [http://dx.doi.org/10.1111/j.1574-6941.2001.tb00890.x doi:10.1111/j.1574-6941.2001.tb00890.x]</ref>. In a SIP study, δ<sup>13</sup>C values for microbial biomass and DIC will be greater than these background levels if biodegradation is occurring because the contaminant degraders are utilizing <sup>13</sup>C from the synthesized contaminant as a carbon and energy source. | The isotopic composition of subsurface microorganisms and their metabolic products (e.g., CO<sub>2 </sub>) will directly reflect the isotopic composition of the compounds used as carbon and energy sources. Thus under natural conditions, background δ<sup>13</sup>C values for microbial biomass and DIC would be expected to be negative and are often between -20‰ and -30‰ <ref>Pelz, O., Chatzinotas, A., Zarda-Hess, A., Abraham, W.R., Zeyer, J., 2001. Tracing toluene-assimilating sulfate-reducing bacteria using <sup>13</sup>C-incorporation in fatty acids and whole-cell hybridization. FEMS Microbiology Ecology, 38(2-3), 123-131. [http://dx.doi.org/10.1111/j.1574-6941.2001.tb00890.x doi:10.1111/j.1574-6941.2001.tb00890.x]</ref>. In a SIP study, δ<sup>13</sup>C values for microbial biomass and DIC will be greater than these background levels if biodegradation is occurring because the contaminant degraders are utilizing <sup>13</sup>C from the synthesized contaminant as a carbon and energy source. | ||
| Line 80: | Line 85: | ||
==Summary== | ==Summary== | ||
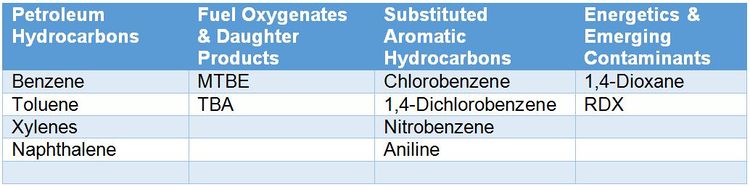
| − | For contaminants that microbes can use as a carbon and energy source (Table 1), stable isotope probing (SIP) is a useful tool for conclusively determining whether in situ biodegradation is occurring. While SIP can aid in evaluating MNA at any site, SIP studies are particularly valuable at complex sites where stakeholders may be reluctant to accept MNA as a site management strategy. Definitive evidence of contaminant biodegradation increases stakeholder confidence that MNA is more than a “do nothing” alternative. At the remedy selection phase, SIP studies can also be performed as part of a pilot study to assess active bioremediation approaches. Greater <sup>13</sup>C incorporation into DIC and biomass in Bio-Traps<sup>®</sup> deployed in wells influenced by site activities (pilot study, injection area, system operation) relative to wells outside of the radius of influence strongly suggests that the remediation technology enhanced biodegradation. In research settings, SIP is used with DNA-based analyses to help identify the organisms involved in specific biodegradation processes<ref name ="Aslett2011"/><ref name = "Hatzinger2014"/><ref name = "Key2013"/>. | + | For contaminants that microbes can use as a carbon and energy source (Table 1), stable isotope probing (SIP) is a useful tool for conclusively determining whether in situ biodegradation is occurring. While SIP can aid in evaluating MNA at any site, SIP studies are particularly valuable at complex sites where stakeholders may be reluctant to accept MNA as a site management strategy. Definitive evidence of contaminant biodegradation increases stakeholder confidence that MNA is more than a “do nothing” alternative. At the remedy selection phase, SIP studies can also be performed as part of a pilot study to assess active bioremediation approaches. Greater <sup>13</sup>C incorporation into DIC and biomass in Bio-Traps<sup>®</sup> deployed in wells influenced by site activities (pilot study, injection area, system operation) relative to wells outside of the radius of influence strongly suggests that the remediation technology enhanced biodegradation. In research settings, SIP is used with DNA-based analyses to help identify the organisms involved in specific biodegradation processes<ref name="Aslett2011" /><ref name="Hatzinger2014" /><ref name="Key2013" />. |
[[File:Table1 SIP Ogles.JPG|thumbnail|center|750px|Table 1. Examples of appropriate contaminants for SIP studies.]] | [[File:Table1 SIP Ogles.JPG|thumbnail|center|750px|Table 1. Examples of appropriate contaminants for SIP studies.]] | ||
| Line 88: | Line 93: | ||
==See Also== | ==See Also== | ||
| + | |||
*[http://www.itrcweb.org/documents/team_emd/SIP_Fact_Sheet.pdf Stable Isotope Probing – Fact Sheet]<br /> | *[http://www.itrcweb.org/documents/team_emd/SIP_Fact_Sheet.pdf Stable Isotope Probing – Fact Sheet]<br /> | ||
*[http://www.itrcweb.org/emd-2/Content/7%20stable%20isotope%20probing%20Sip.htm Stable Isotope Probing (SIP)]<br /> | *[http://www.itrcweb.org/emd-2/Content/7%20stable%20isotope%20probing%20Sip.htm Stable Isotope Probing (SIP)]<br /> | ||
| − | *[https://www.serdp-estcp.org/Program-Areas/Environmental-Restoration/Contaminants-on-Ranges/Protecting-Groundwater-Resources/ER-1606 Development of Biomarkers for Assessing In | + | *[https://www.serdp-estcp.org/Program-Areas/Environmental-Restoration/Contaminants-on-Ranges/Protecting-Groundwater-Resources/ER-1606 Development of Biomarkers for Assessing In Situ RDX Biodegradation Potential ER-1606]<br /> |
*[https://www.serdp-estcp.org/Program-Areas/Environmental-Restoration/Contaminants-on-Ranges/Characterizing-Fate-and-Transport/ER-1607/ER-1607 New Approaches to Evaluate the Biological Degradation of RDX in Groundwater ER-1607]<br /> | *[https://www.serdp-estcp.org/Program-Areas/Environmental-Restoration/Contaminants-on-Ranges/Characterizing-Fate-and-Transport/ER-1607/ER-1607 New Approaches to Evaluate the Biological Degradation of RDX in Groundwater ER-1607]<br /> | ||
*[[Characterization, Assessment & Monitoring]]<br /> | *[[Characterization, Assessment & Monitoring]]<br /> | ||
| − | *[https://www.coursera.org/learn/natural-attenuation-of-groundwater-contaminants/lecture/83aUM/stable-isotope-probing | + | *[https://www.coursera.org/learn/natural-attenuation-of-groundwater-contaminants/lecture/83aUM/stable-isotope-probing Online Lecture Course - Stable Isotope Probing] |
Latest revision as of 22:08, 26 April 2022
Stable isotope probing (SIP) uses heavy isotopes to identify and track contaminant fate to evaluate whether biodegradation is occurring at a site. Here, we describe how SIP works in a typical study, outline its advantages and limitations, briefly walk through reporting and background values, introduce a case study use at a petroleum hydrocarbon site, and summarize suggested guidelines for use in other studies.
Related Article(s):
- Compound Specific Isotope Analysis (CSIA)
- Metagenomics
- Molecular Biological Tools - MBTs
- Quantitative Polymerase Chain Reaction (qPCR)
Contributor(s): Dora Ogles-Taggart and Dr. Brett Baldwin
Key Resource(s):
- Identification of TBA-utilizing organisms in BioGAC reactors using 13C-DNA SIP[1]
- Using DNA‐Stable Isotope Probing to Identify MTBE‐and TBA‐Degrading Microorganisms in Contaminated Groundwater[2]
Introduction
Stable isotope probing (SIP) is used to conclusively determine whether in situ biodegradation of a contaminant is occurring. The “probe” is a synthesized version of the contaminant compound composed of the heavier stable isotope (e.g., 13C, 15N) rather than the more common light isotope of that element (e.g., 12C, 14N). The heavy isotope serves as the “label” to track the environmental fate of the contaminant and determine if biodegradation is occurring.
SIP is used to conclusively determine if biodegradation of some common contaminants like petroleum hydrocarbons (e.g., BTEX, polycyclic aromatic hydrocarbons (PAHs)) and oxygenates (e.g., MTBE, TBA) is occurring. Results are often used to evaluate the feasibility of monitored natural attenuation (MNA) as a site management strategy[3][4][5][6]. Conclusive evidence of contaminant biodegradation increases stakeholder confidence that MNA is more than a “do nothing” alternative. While often performed to assess MNA, SIP can also be used to evaluate the feasibility and performance of engineered bioremediation approaches. In research settings, SIP can also be used with DNA-based analyses to help identify the organisms involved in specific biodegradation processes[1][7][2].
How Does SIP Work
In a typical SIP study, a 13C form of the contaminant (e.g., 13C-benzene) is adsorbed to a passive microbial sampling device such as Bio-Sep® beads inside a Bio-Trap® sampler. Bio-Sep® beads are an engineered composite of Nomex® and powdered activated carbon (PAC). PAC adsorbs the 13C-labeled compound and also provides a large surface area for microbial colonization and growth. Nomex® allows the beads to be heat sterilized prior to the study. Such a passive sampler is deployed in a monitoring well (Fig. 1).
During the deployment period (30 to 90 days), the 13C-labeled contaminant is subject to the same microbial processes as unlabeled contaminant present at the site. Many contaminants, such as petroleum hydrocarbons, are used as a carbon and energy source for microbial growth. Therefore, if biodegradation is occurring, contaminant-degrading bacteria will colonize the Bio-Trap® and use the 13C-labeled contaminant as a carbon and energy source for growth and the 13C label will be incorporated into microbial biomass or 13CO2.
After deployment, the Bio-Trap® is recovered for gas chromatography and isotope ratio mass spectrometry (IRMS) analysis to quantify the 13C/12C ratio of biomass and dissolved inorganic carbon. The 13C label will either be incorporated into microbial biomass or mineralized to 13CO2. Detection of 13C-enriched biomolecules (phospholipids, DNA, or protein) and 13C-enriched dissolved inorganic carbon (DIC) following deployment unambiguously indicates that in situ biodegradation occurred. Conversely, if biodegradation is not occurring, the 13C/12C ratio of the microbial biomass and DIC analyzed after in well deployment will be similar to background values. Phospholipid fatty acids (PLFA) are a main component of microbial cell membranes, therefore, 13C-enriched PLFA unambiguously demonstrates incorporation of the 13C into biomass. Likewise, 13C-enriched dissolved inorganic carbon (e.g., CO2 , HCO3-) provides conclusive evidence of contaminant mineralization.
Advantages
- Conclusive: SIP can provide conclusive evidence that in situ biodegradation of the contaminant is occurring.
- Broadly applicable: A SIP study can be conducted for any contaminant that is used as a carbon and energy source, as long as an isotopically-enriched form of the contaminant can be synthesized.
- No prior knowledge needed: No prior knowledge is needed about the type(s) of microorganisms, biodegradation pathway(s), or gene sequences.
- Reasonable cost: For many common contaminants like BTEX, MTBE, TBA, and even naphthalene, the cost to synthesize the 13C-labeled compound is reasonable. As an example, the cost for a SIP study with 13C benzene including the cost of synthesizing the 13C compound, post-deployment analysis, and reporting is ~ $1,000.
Limitations
- Not applicable to all contaminants: SIP is generally not appropriate for compounds that are used as terminal electron acceptors, such as trichloroethylene (TCE) and other chlorinated ethenes, because the 13C label is not incorporated into biomass or CO2 during this microbial process. Tools such as compound-specific isotope analysis (CSIA) performed on the contaminant itself is more applicable for these compounds. For large or more complex compounds, synthesis of the 13C-labeled compound can be expensive or simply not available.
- Dilute plumes: Data obtained from typical SIP studies where concentrations of 13C-labeled compounds are relatively high may not necessarily extrapolate to biodegradation of the contaminant present in a dilute plume where contaminant concentrations are approaching closure levels.
Reporting SIP Results
SIP results are reported as 13C/12C ratios in delta notation (δ13C) with units of parts per thousand (“per mil” ‰), where δ13C is defined as follows[8][9]:
- Where:
- Rsample is the 13C/12C ratio of the sample and
- Rstandard is the 13C/12C ratio of the Vienna Pee Dee Belemnite (VPDB) standard.
Background δ13C Values
The 13C/12C ratio of the VPDB standard (1.1112328% 13C) is greater than most other natural carbon-based substances[8]. Because Rstandard is typically larger than Rsample , calculated δ13C values of most naturally-occurring materials will be negative values (0‰ to -110‰).
The isotopic composition of subsurface microorganisms and their metabolic products (e.g., CO2 ) will directly reflect the isotopic composition of the compounds used as carbon and energy sources. Thus under natural conditions, background δ13C values for microbial biomass and DIC would be expected to be negative and are often between -20‰ and -30‰ [10]. In a SIP study, δ13C values for microbial biomass and DIC will be greater than these background levels if biodegradation is occurring because the contaminant degraders are utilizing 13C from the synthesized contaminant as a carbon and energy source.
Use for Evaluating MNA at Petroleum Hydrocarbon Sites
At complex petroleum hydrocarbon sites, observing decreasing trends in contaminant concentrations and plume stability may be insufficient to conclusively evaluate the feasibility and performance of MNA. SIP can provide a critical third line of evidence to conclusively determine if biodegradation of a specific contaminant is occurring under the existing subsurface conditions and improve stakeholder confidence in MNA as a site management strategy.
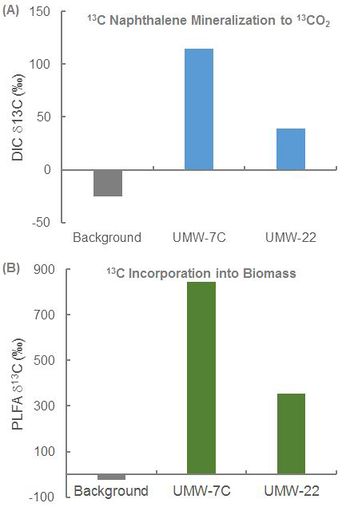
In an ongoing study, SIP was conducted at a former manufactured gas plant (MGP) to determine whether naphthalene biodegradation was occurring under existing site conditions and whether MNA would be effective. Bio-Traps® amended with 13C-naphthalene were suspended in two monitoring wells (UMW-7C and UMW-22) located within the dissolved plume for approximately 60 days. After the deployment period, the Bio-Traps® were recovered for analysis.
- Detection of 13C-enriched DIC in Bio-Traps® recovered from UMW-7C and UMW-22 demonstrated mineralization of 13C naphthalene under existing site conditions (Fig. 2A).
- Detection of 13C-enriched PLFA demonstrated 13C incorporation into biomass (Fig. 2B).
Overall, the SIP study conclusively demonstrated biodegradation of naphthalene under the existing site conditions. This was a critical line of evidence in support of an MNA remedy.
SIP Study Guidelines
As with any site assessment tool, study design and proper implementation are needed to ensure the relevance and accuracy of the results. Here are some suggestions:
- Study locations: SIP studies with a 13C labeled contaminant should only be performed in impacted areas within the dissolved plume. Comparisons of results to literature δ13C values for DIC and PLFA provide meaningful conclusions. If desired, site specific δ13C values for DIC and PLFA can be determined from water samples or standard Bio-Traps® (not amended with a 13C compound).
- Deployment period: Typical deployment periods are between 30 and 90 days although longer deployment periods have been used to investigate biodegradation of more recalcitrant compounds.
- Handling: Clean latex gloves (or similar) should be worn at all times when handling Bio-Traps®. Bio-Trap® samplers should be kept in sealed bags and refrigerated, but not frozen until deployment. After deployment, Bio-Traps® should be bagged and immediately placed on ice. Samplers should be shipped on ice using an overnight carrier.
Summary
For contaminants that microbes can use as a carbon and energy source (Table 1), stable isotope probing (SIP) is a useful tool for conclusively determining whether in situ biodegradation is occurring. While SIP can aid in evaluating MNA at any site, SIP studies are particularly valuable at complex sites where stakeholders may be reluctant to accept MNA as a site management strategy. Definitive evidence of contaminant biodegradation increases stakeholder confidence that MNA is more than a “do nothing” alternative. At the remedy selection phase, SIP studies can also be performed as part of a pilot study to assess active bioremediation approaches. Greater 13C incorporation into DIC and biomass in Bio-Traps® deployed in wells influenced by site activities (pilot study, injection area, system operation) relative to wells outside of the radius of influence strongly suggests that the remediation technology enhanced biodegradation. In research settings, SIP is used with DNA-based analyses to help identify the organisms involved in specific biodegradation processes[1][7][2].
References
- ^ 1.0 1.1 1.2 Aslett, D., Haas, J., Hyman, M., 2011. Identification of tertiary butyl alcohol(TBA)-utilizing organisms in BioGAC reactors using 13C-DNA stable isotope probing. Biodegradation, 22(5), 961-972. doi:10.1007/s10532-011-9455-3
- ^ 2.0 2.1 2.2 Key, K.C., Sublette, K.L., Duncan, K., Mackay, D.M., Scow, K.M., Ogles, D., 2013. Using DNA‐Stable Isotope Probing to Identify MTBE‐and TBA‐Degrading Microorganisms in Contaminated Groundwater. Groundwater Monitoring & Remediation, 33(4), 57-68. doi:10.1111/gwmr.12031
- ^ Busch‐Harris, J., Sublette, K., Roberts, K.P., Landrum, C., Peacock, A.D., Davis, G., Ogles, D., Holmes, W.E., Harris, D., Ota, C., Yang, X., 2008. Bio‐Traps Coupled with Molecular Biological Methods and Stable Isotope Probing Demonstrate the In Situ Biodegradation Potential of MTBE and TBA in Gasoline‐Contaminated Aquifers. Groundwater Monitoring & Remediation, 28(4), 47-62. doi:10.1111/j.1745-6592.2008.00216.x
- ^ Geyer, R., Peacock, A.D., Miltner, A., Richnow, H.H., White, D.C., Sublette, K.L., Kästner, M., 2005. In situ assessment of biodegradation potential using biotraps amended with 13C-labeled benzene or toluene. Environmental Science & Technology, 39(13), 4983-4989. doi:10.1021/es048037x
- ^ Key, K.C., Sublette, K.L., Johannes, T.W., Ogles, D., Baldwin, B., Biernacki, A., 2014. Assessing BTEX Biodegradation Potential at a Refinery Using Molecular Biological Tools. Groundwater Monitoring & Remediation, 34(1), 35-48. doi:10.1111/gwmr.12037
- ^ Williams, N., Hyland, A., Mitchener, R., Sublette, K., Key, K.C., Davis, G., Ogles, D., Baldwin, B., Biernacki, A., 2013. Demonstrating the In Situ Biodegradation Potential of Phenol Using Bio‐Sep® Bio‐Traps® and Stable Isotope Probing. Remediation Journal, 23(1), 7-22. doi:10.1002/rem.21335
- ^ 7.0 7.1 Hatzinger, P., Fuller, M., 2014. New approaches to evaluate the biological degradation of RDX in groundwater. Project ER-1607. Strategic Environmental Research Development Program, Arlington, VA. ER-1607
- ^ 8.0 8.1 Laboratory S.I.E. Overview of Stable Isotope Research. http://sisbl.uga.edu/stable.html
- ^ USGS Fundamentals of Stable Isotope Geochemistry. USGS Resources on Isotopes
- ^ Pelz, O., Chatzinotas, A., Zarda-Hess, A., Abraham, W.R., Zeyer, J., 2001. Tracing toluene-assimilating sulfate-reducing bacteria using 13C-incorporation in fatty acids and whole-cell hybridization. FEMS Microbiology Ecology, 38(2-3), 123-131. doi:10.1111/j.1574-6941.2001.tb00890.x
See Also
- Stable Isotope Probing – Fact Sheet
- Stable Isotope Probing (SIP)
- Development of Biomarkers for Assessing In Situ RDX Biodegradation Potential ER-1606
- New Approaches to Evaluate the Biological Degradation of RDX in Groundwater ER-1607
- Characterization, Assessment & Monitoring
- Online Lecture Course - Stable Isotope Probing